Solve your Y-Chromosome Puzzle with Yfull!
Yfull is not a provider of Y-DNA or NGS tests, but an analysis service for NGS with a database in which data from NGS tests are compared and analyzed, regardless of the provider. This allows to compare the results of currently 13 sources. (Commercial companies and scientific studies) See: phylogeographer.com/yfull-cost-benefit-analysis. The results are presented in the form of a Y-tree in different representations. The kits are presented anonymously and the Y-tree (as well as the mt-tree) is publicly accessible for everyone. This makes it not only an enrichment for the submitters, but also for everyone who is concerned with the topic Y-tree (as well as mt-tree).
1.0 Ordering the analysis
- Preparation: You don’t upload the raw data of the NGS test to Yfull yourself, but indicate where it can be downloaded. You can get the link from the provider where you did your test. (FTDNA, YSEQ, FGC). Companies like Dante Labs don’t offer such a link for Yfull directly, so you have to make the required file accessible via a cloud. You can find an easy instruction here: Prepare the BAM file for YFull.
- Go to the Yfull site and order the analysis for $49: https://www.yfull.com/order/
- The selection of VCF as raw data file type is only possible for FTDNA BigY. The preferred raw data file type is BAM. For other companies only this can be selected.
- Specify the testing company.
- Enter the URL to your BAM file.
- Under “Comment” you can send additional information to the Yfull Team. This is especially important if you send several kits from the same participant. (e.g. This is an upgrade of BigY-500 kit YF10815)
- Order Now!
You will receive a confirmation email immediately:
Hello! Your order will be verified by the manager. If the raw data link is correct, your analysis will be batched. Login and password for new clients to access the site will be sent. Notification for payment will be sent after completion of the interpretation. Thanks.
2.0 Analysis of the raw data – a time line
2.1 The SNP Analysis – Donated Kits
Now we have to wait. If the link works, the analysis is batched and you get access data and the “YF number” for the kit. This can take some hours, but also a few days. The payment, which unlocks all functions, will be done *after* the analysis.
Some days later the kit is placed in the Y-tree with a temporary position. The YF number is grey and a small box with “new” is located behind the kit. The exact position is only obtained after the analysis and the update of the tree (the analysis takes about four weeks).
2.1.1 Settings:
Now, you can and should make some settings. (Button “Settings”, top right).
Account Settings:
- COUNTRY OF ORIGIN = place of origin of the most distant known ancestor in the paternal line (Tab Y).
- MOST DISTANT ANCESTOR = most distant known ancestor in the paternal line (Tab Y).
- USERNAME = Name of the user or nickname for this kit. This information makes it easier to choose if you administer several kits or participate in groups.
Sharing Settings:
Here you can share the results with Yfull users. This is especially interesting if you want a friend to explain the results to you. To share the results with another Yfull user, send an invitation to the email address with which the participant is registered at Yfull.
2.1.2 Useable functions at this stage:
Two functions can be used while waiting for the results. “Browse raw data” and “Statistics”.
With “Browse raw data” you can examine the raw data and query individual ChrY and ChrM positions. With some practice and knowledge of the novel variants of the other participants, you can determine your terminal SNP. (This function is comparable to function 3.1.4 Check SNPs)
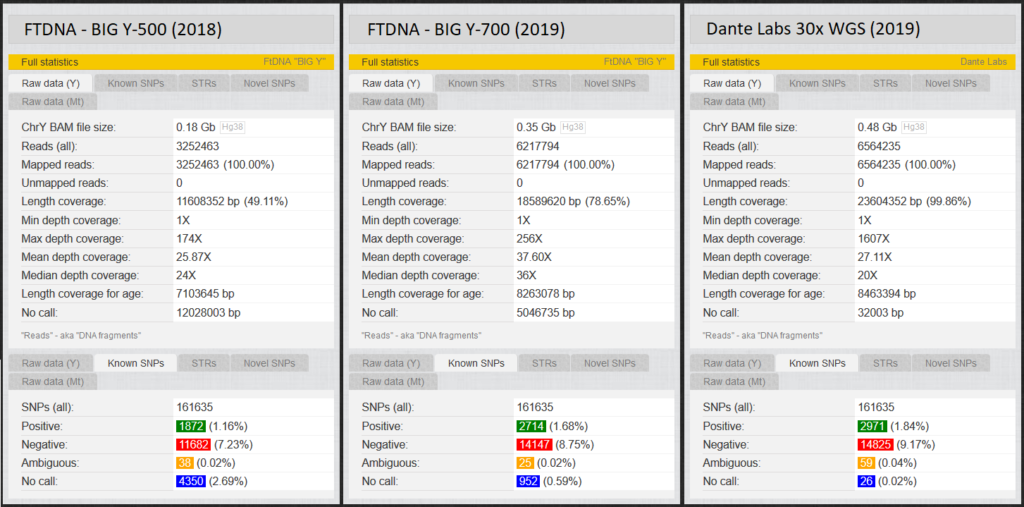
On “Statistics” you get a small statistic regarding the quality of the raw data. The figure shows the statistics of two men (TMRCA 50 ybp). Big Y-500, same man upgrade to Big Y-700, fifth degree cousin WGS at Dante Labs.
2.1.3 Unlock results, or “Donate Kit“.
After about four weeks the first part of the analysis is finished and you can have the results unlocked with payment. There is also the possibility to “donate” a kit. Here only the SNP analysis is performed, without STR analysis, and the kit does not participate in the age estimation. I personally see no reason to do this, as I am interested in the complete analysis. However, it is a benefit for the Y-tree and the projects if people with limited interest (e.g. WGS testers with focus on health) submit their kits that way.
2.2 After unlocking: STR analysis and age estimation
After paying the one-time fee of $49, all functions are unlocked and the STR analysis and age estimation is initiated. The STR analysis will take a few days, or weeks. The age determination is done just before the next update of the tree, so this may take a few weeks. All other functions are available immediately, but you should be aware that there will be some changes regarding the new SNPs until the next update of the tree.
2.2.1 Uploading STR values
If you have performed a Y-STR test in addition to the NGS test, for FTDNA or YSEQ, you can upload the STR values. https://www.yfull.com/ls-upload-strs/ STR values that cannot be extracted from the NGS raw data are added in this way.
2.2.2 Uploading the MT FASTA file
A WGS contains the entire mtDNA in addition to the entire Y chromosome, so that mt is added to the Yfull mt tree. If you participated with an NGS without mtDNA, you can upload your FASTA file additionally. https://www.yfull.com/mt-upload-list/
You can and should now make some settings. (Button “Settings”, top right).
Account Settings:
- COUNTRY OF ORIGIN = place of origin of the most distant known ancestor in maternal line (tab mtDNA)
- MOST DISTANT ANCESTOR = most distant known ancestor in maternal line (tab mtDNA).
- USERNAME = Name of the user or nickname for this kit. This information makes it easier to choose if you administer several kits or participate in groups.
2.2.3 Join YFull Groups
Check out the list of Yfull groups at https://www.yfull.com/groups/list/ and join the group that matches your haplogroup.
2.3 Positioning in the Y-tree
You only get a preliminary position. After successful SNP analysis and update of the tree you get the next “Preliminary” position. Which positions you can take has already been described in NGS – Take part in the Y-Tree. The final position in the Y-tree is “never” obtained, because the Y-tree “lives”. With each additional kit on the relevant clades, the shape of the tree can change.
3.0 YFull functions
It is worth hoovering the mouse pointer over the Yfull interface and experiencing for yourself what information and links are connected behind the buttons.
3.1 SNPs
3.1.1 Hg and SNPs
This lists all SNPs that have tested positive, are likely to be positive, or are No Calls. The green box shows your terminal SNP, or the name of the block of your terminal SNPs (here I-Y158862). Below it, all terminal SNPs of this block are listed in sequence (here A23501/BY182855 and BY182587/Y158862).

Under “Known SNPs”> Positive, all SNPs are listed that have tested positive with this kit and are “known”. These are the SNPs that have been found in other kits before. Yfull evaluates the quality of these SNPs with an internal star system.
Figure 2
shows what this view looks like just before new branches are formed.
Y158878 • BY183648, level I-Y158878, terminal new, five stars.
- Y158878 and BY183648 are two different names for the same SNP.
- Level I-Y158862 indicates the block to which this SNP (still) belongs.
- “Terminal new” indicates that something is going on. These SNPs have only recently been shared with another kit and were previously under “Novel SNPs”. After the next update of the tree, these SNPs will form the new block of terminal SNPs.
- Only the upper two SNPs have five stars rating. One of these two SNPs will probably be used to label this new block.
- SNPs with the addition “private” are mostly low quality SNPs that have already been found in several kits, before.
- If you click on the magnifying glass in front of the SNPs, you will see a view with more information about the respective SNP in a kit. (see 3.1.4)
3.1.2 Novel SNPs
The Novel SNPs are SNPs that are only found in your kit. They are divided into five categories. “Best qual” and “Acceptable qual” have very good or sufficient quality. “Ambigous qual” and “Low qual” have only moderate quality. You cannot be sure that these are actually positive, because they have either not been read often enough or do not show clear results. It is recommended to make them orderable at YSEQ with “Wish a SNP” and to check them for correctness. More about this topic here: Verify relationship in paternal line, with known terminal SNP and private „novel“ SNPs from NGS
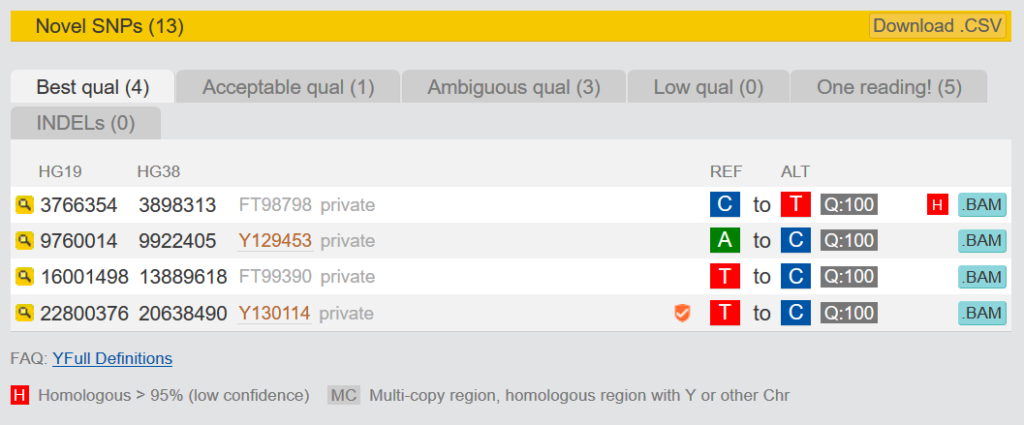
SNPs that can be ordered at YSEQ are marked with a small sign at Yfull. If this sign appears orange, this has never been tested positive, before. A green sign stands for SNPs that have already tested positive. Hoover the mouse over this sign to see how often it has been tested.
If you click on the magnifying glass in front of the SNPs, you will see a view with more information about the respective SNP in a kit. (see 3.1.4)
Clicking on the blue “.BAM” field opens a Y-Browser.
3.1.3 SNP Matches
This view shows the SNP matches as they are also shown on the Y-tree, but you get some more information about the user. Firstly, the “Most Distant Ancestor” and secondly, the information which SNPs you actually share and which SNPs you only ambiguously share.
3.1.4 Check SNPs
This function is similar to the previously mentioned “Browse raw data”. However, here you can enter the name of the SNPs. On the right hand side you can see immediately whether the SNP has been tested positive or negative. (The green sign behind I-L38 stands for “Verified by Sanger Sequencing, YSEQ tested”).
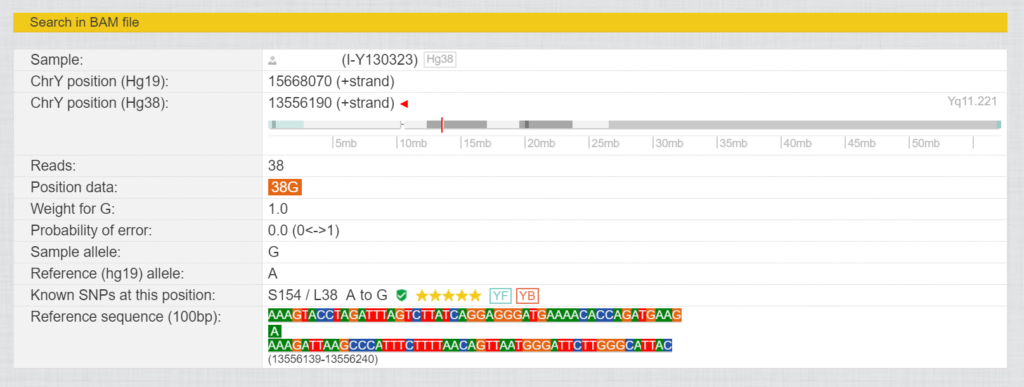
If you click on the magnifying glass in front of the SNPs, you will see a view with more information about the respective SNP in a kit.
Here you can find out the ChrY position (Hg19) and (Hg38) for this SNP, as well as in which area of the Y chromosome it is located. “Reads” indicates how often this was read.
The ChrY position (Hg38) for L38 is 13556190 in the region of Yq11.221. This was read 38 times in this kit and resulted in 38 times “G”. The reference has “A” at this point. The known SNPs at this position are S154/L38 from A to G, “Verified by Sanger Sequencing, YSEQ tested”, 5 star rating, YF= in Yfull database, YB=in ISOGG YBrowse database.
Reference sequence (100bp) shows which values the reference has 50 bases before and 49 bases after this position.
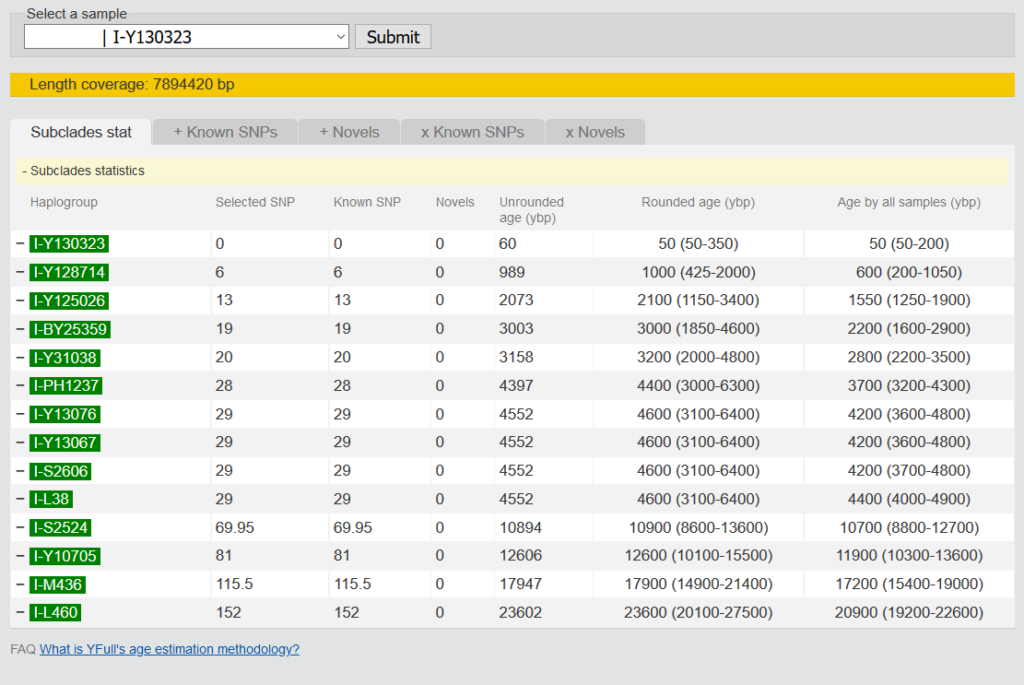
3.1.5 Age Estimation
The age estimation of a clade is based on the positive SNPs of all samples. On the overview you can see the influence of your own kit on the age estimation of all samples. In the other tabs you can see which SNPs were taken for the age estimation and which ones were not taken and why. A detailed description of the method can be found by following the link below the table. What is YFull’s age estimation methodology?
3.1.6 Upgrades
There are two upgrade options. This applies *only* to differently prepared raw data of the same test. (An “upgrade” from Big Y-500 to Big Y-700 is not an upgrade, but a new test)
- Upgrade the analysis of a Big Y, from raw data format VCF to BAM. This upgrade is free of charge and is recommended, as only the BAM contains all information for a comprehensive analysis.
- Upgrade of the analysis from a NGS, from Hg19 to Hg38. This upgrade is possible, but Yfull does not recommend it.
For each upgrade you do, you get a comparison of the results for:
- Known SNPs
- Novel SNPs
- STRs
- Statistics
3.1.7 Comparisons
If a person has performed several tests, e.g. Big Y-500 vs. Big Y-700 or Big Y-500 vs. WGS YSEQ, the results can be compared here. These are:
- Known SNPs
- Novel SNPs
- STRs
- Statistics
3.1.8 YReport
This is an overview of all positive SNPs of a kit from the Y-Adam, up to the tester. You can see at a glance which SNPs have been read, which are No Calls or “Ambigous”. Selecting the SNP shows us the window, which we already know from 3.1.4, with the values for this SNP.
3.2 STRs
The raw data from NGS not only contain information on SNPs. Yfull can read out up to 780 STRs and offers some nice additional features.
3.2.1 STR results
The 780 STRs are listed here. The different colours of the boxes, stand for:
- White = reliable STR
- Grey = uncertain STR
- Blue = STR taken from YSEQ or FTDNA file
You can download this file as .CSV.
3.2.2 STR matches
Matches based on STRs are listed here. For this purpose, the STRs that differ in the kits are simply summed up. About sense and nonsense of this representation one can argue. I do not find it informative.
3.2.3 STR Variants
However, this function is very informative. Here you get the mutation course of your STRs from Y-Adam to yourself. They are sorted from “young” to “old”, linked to the information within which SNP blocks they are mutated.

For a better overview, the figure shows only the selection for Y111. The first column shows the label of the SNP block in which the STR is mutated. Next to the STR you get the information which other kits share this STR with you. On the right you can see values for:
- The detection rate – In which part of all kits was this STR detected?
- The mutation rate – Five stars stand for an STR which rarely mutates. The fewer stars, the more likely this STR tends to mutate, so that two kits can randomly have the same value.
- Ancestral (ANC) and derived (DER) value for this STR.
This view can help you to understand the order of STR mutations. Under certain circumstances, subgroups can be formed based on STR values, where no distinction is possible with SNPs. However, caution is advised, especially if the mutation rate is low.
STRs with a mutation rate of five stars and sufficient samples are actually used to form subgroups with YFull.
3.3 Contacting YFull users
If you are a user of Yfull you can contact any other user. After clicking on “New message”, simply enter the number after “YF” as “Recepient” (arrow 3).
4.0 Groups at Yfull
Check out the list of Yfull groups at https://www.yfull.com/groups/list/ and join the group that matches your haplogroup. This allows you and the administrators of the group to conduct research within the group.
4.1 Y-Browser

With the Y-Browser you can compare values for individual Y-Chr HG38 positions of the group users. You can also compare positions like “Novel Variants”, which is not possible with the “View SNPs” of “Y-Results”. This way you can see at once if there are test persons who have no calls for these positions and could theoretically be positive for these new SNPs. In case of increased suspicion (e.g. common STR mutations) the new SNPs can be tested on these test persons after wishing the SNPs with “Wish a SNP” for being orderable.
4.2 Y-Results
4.2.1 Y-STR Group viewer
With “View Y-STRs (classic)” and “View Y-STRs (coloured)” you get a table with the 780 STRs of all group members.
- “View Y-STRs (classic)” shows a classic view. It contains the information about the reliability with which a STR (reliable or uncertain) was extracted.
- “View Y-STRs (coloured)” shows the modal values, minimum values and maximum values in colour. Here the information regarding the reliability of the STRs is missing.
- This table can be downloaded and used for STR investigations.

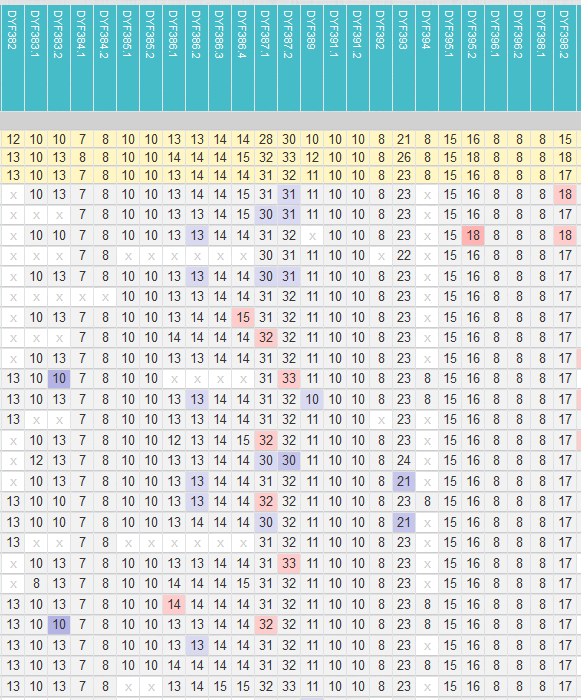
4.2.2 SNPs Group viewer
This function is similar to the Y-Browser, but has the advantage that you can enter the SNPs directly. Therefore you cannot select “New SNPs” here. You can immediately see which SNPs have *not* been tested with which testers (No Calls). So you can find potential candidates for not yet discovered subgroups.
In the example in Figure 12, the SNPs of block Y177573 were queried. Most of the SNPs were read out for all participants. SNPs Y177575 were only read in two kits with WGS and one Big Y-700, out of a total of four Big Y-700. SNP Y177577 has only been read by two WGS (green and red arrow). For the participants with Big Y these are No Calls. One could now make these two SNPs orderable at YSEQ with “Wish a SNP” and then check if the kits of Y125026* are positive for them.

This is also one of the reasons why I prefer a WGS in comparison to a targeting NGS like Big Y. The better coverage and therefore hardly any no calls. (see fig. 1)

Pingback: Yfull - Sign In - Yfull
Pingback: yfull login - LoginWhale.com
Hello! I am Paula Paz. I am a researcher with expertise in Y-Chromosome analysis and I am eager to continue my career with the entire YFull team. Is there a specific email address I can use to send my application and CV? Thank you very much!
Try acgt@yahoo.com