Entry, tests for DNA
I want to test my DNA. Which test should I take?
This question is often read in this or a similar way in various forums, but without a crystal ball it will be difficult to find an answer, if you do not know the intentions of the questioner. Only those with sufficient information can be answered accurately. First of all, the questioner should be clear about what he wants and formulate it correctly. Basic knowledge about the topic facilitates this. This article is intended to provide an initial introduction to the topic, roughly show what intentions and methods there are and make it easier for test takers to ask questions and communicate.
What can be tested and why?
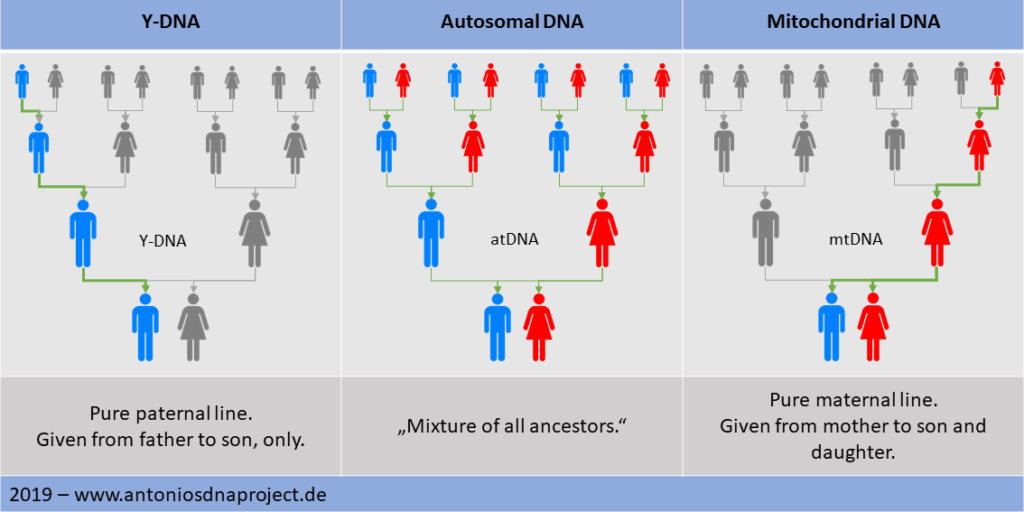
Autosomal DNA:
Most testers are
available for autosomal DNA tests. Accordingly, there are some companies with
large user databases. AtDNA tests are very often advertised with the “ethnicity
estimation” and are therefore bestsellers. Unfortunately, they often have a bad
reputation because of this, because critical articles often don’t deal with the
strengths of these tests at all or hardly at all.
Autosomal DNA can be very helpful for genealogy, because you can do the following with it, also because of the large user databases:
- Verification of single ancestor lines of the family tree,
- Closing gaps in the family tree (e.g. finding out who the grandfather or great-grandfather was)
- Identify the biological parents,
- Finding distant relatives who have emigrated (or who have not) and contacting them.
Mitochondrial DNA:
Mitochondrial DNA is passed on from the mother to sons and daughters and allows
conclusions to be drawn about the relationship of the pure female line. It
mutates very slowly, so that it remains unchanged even after many generations.
This means that it will only be useful for genealogy in special cases, which
does not mean that it cannot provide valuable information, especially in
migration pathways based on mt-haplogroups.
Y-DNA:
This is at the
heart of this report and will receive the most attention here. Nevertheless, I
would like to use a small example to illustrate how important it is to first
think about exactly what one is planning to do, to devise a strategy, and to
think about which tool (or tools) is best suited to achieving one’s goal. By
communicating strategy and plans, you can get valuable tips and optimize them.
One is quickly inclined to want to process everything related to the paternal line with the Y-DNA tool, even if there is an easier possibility. Let’s take the previously mentioned case: “Closing gaps in the family tree and finding the paternal grandfather”. This may work with Y-DNA in certain cases, but the better tool for this is the atDNA, just because of the much larger database and the possibility to find the grandfather by side lines.
But when it comes to verifying paternal relationships over several generations, the Y-DNA test is the right choice. My own concern is to verify a family legend that says that my fatherly ancestor came from a certain area of Greece about three hundred years ago. Here you can’t get any further with atDNA. The selection of tests for Y-DNA is extensive. Depending on the motivation for such a test, there are different approaches.
Y-DNA, Methods and Intentions
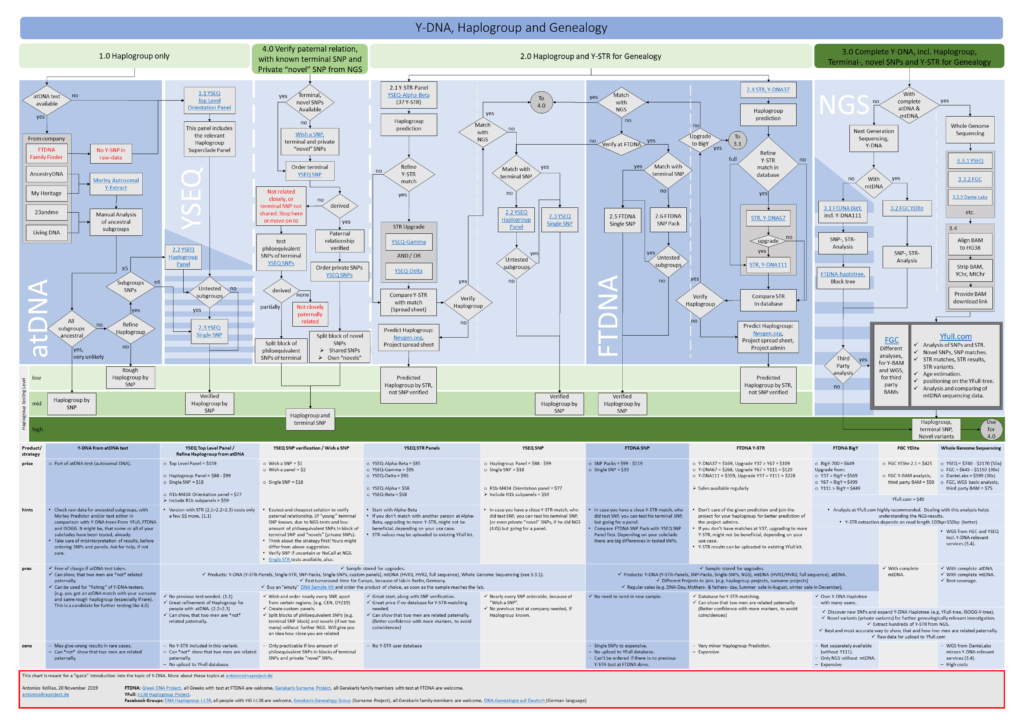
This flow chart can be downloaded here: Y-DNA, Haplogroup and Genealogy.PDF
1.0 Haplogroup only
Not everyone who thinks of a test for Y-DNA has the genealogy and his direct paternal ancestors in mind. Some are “only” interested in the Y-haplogroup for various reasons, such as historical migrations. In the autosomal DNA tests of 23andme and Living DNA, the Y-haplogroup and the mt-haplogroup are displayed directly. What many people don’t know is that all autosomal tests include information in the form of SNPs (Single Nucleotide Polymorphism) for the Y-haplogroup and are present in the raw data. Only FTDNA removes this information from the raw data. With a script called Morley Predictor this information can be extracted.
It is often overlooked that not only the Y-haplogroup, which has been determined, is important, but also the subgroups, which have already been tested as negative. Unfortunately, this is not displayed directly. If you get an “old” haplogroup displayed (23andme, Living DNA), although younger branches are also tested, it can mean that you are sitting on a rare branch. A manual check of the raw data is advisable.
Different providers test different numbers of Y-SNPs. Nevertheless, I maintain that the results are at almost the same low level. If you want to know your Y-Haplogroup more exactly and find out to which subgroup you belong, you have to use a so-called „Haplogroup panel“ or single SNPs. The easiest way to do this is with the company YSEQ from Berlin, without having to take an expensive detour via Y-STR (Short Tandem Repeat). If you already know your rough haplogroup, you can climb down the Y-tree to your youngest known (terminal) SNP, for little money. If you haven’t done an atDNA test before and aren’t interested in their advantages, you can directly choose the „Top Level Orientation Panel“, which leads to the same result, a haplogroup verified by SNP.
What is a Y-haplogroup?
A Y-haplogroup is a
group of men who share a common ancestor in a paternal line. This is called
MRCA for „Most Recent Common Ancestor“. If you talk about the time, how
far back he lived, you talk about TMRCA for “Time to the Most Recent
Common Ancestor”. With a simple determination of the Y-haplogroup, the
TMRCA can be about 5000 years or far beyond, and even if you refine to
subgroups, you may not get closer than 1000 years to the MRCA. If you are
concerned with genealogy, it can also be interesting how your Y-haplogroup has
spread over the last 2000 years, but the main interest might be in the last
centuries. For this we need methods to look at the mutations of the last
centuries.
Summary, intentions for (1.0):
- Determination of the Y-haplogroup for prehistoric research.
- While these tests do not indicate that two men are related in the paternal line, they do indicate that two men are *not* related in the paternal line, which can often be helpful.
- Due to the large databases and the associated autosomal matches, this can also be used well for “fishing”, i.e. finding testers who are relevant to one’s own Y-research and encourage them to take a further test.
2.0 Haplogroup and Y-STR for genealogy
One of these methods, Y-STR (Short Tandem Repeat), is relative old but still common practice. A few years ago, there were still several companies using Y-STR, including AncestryDNA, but in the meantime only a few companies still offer such Y-STR panels. Two notable companies are FamilyTreeDNA (FTDNA) in Texas, Houston and YSEQ in Berlin, Germany. If you want to do a test for Y-DNA at FTDNA, you have to buy at least Y-37. (Smaller panels like Y12 and Y25 can only be purchased through projects.) The equivalent product at YSEQ is called YSEQ-Alpha-Beta.
The basic question, which of these two companies should be used for a Y-STR test, is not easy to answer. FTDNA’s panels are more expensive, but they have a user database and project groups that you can join. Whether you benefit from the database you can’t know beforehand, but only after testing. As a rule, the more testers there are in your region of origin, the more likely you are to benefit from this database, but there are likely to be some exceptions, so I don’t want to make a recommendation here.
The clear advantage of Y-STR is that you can compare the Y-DNA of two or more men in a database or spread sheet, since Y-STR can also contain younger mutations. The more Y-STR markers are available, the more accurate the conclusions can be about the degree of kinship. If you don’t have anyone to compare with (no match) when starting with 37 markers, it is questionable whether more markers is useful and it makes sense to expand to Y67 or Y111. This depends on several factors and your own motives.
Y-haplogroups are defined via Y-SNP and named after them. But they can be, more or less “predicted” via Y-STR. Such a prediction is shown at your FTDNA Y-results tab, if you do just a Y-STR test but this is very conservative, sometimes so primitive that one wonders why one has spent so much money on it. This is because you can only *predict* the Y-Haplogroup by Y-STR, but the verification is done by Y-SNP. Depending on the number of Y-STR, the haplogroup and the ability of an expert to interpret this haplogroup (haplogroup administrators), you can even make more accurate predictions than you can verify with SNP packs. In the same way it can happen that a predestination is not possible. A useful tool to predict the haplogroup from the Y-STR is the Nevgen Predictor.
In many cases it is necessary to verify the predicted haplogroup via SNP. Here you should think about whether it makes more sense to take an SNP pack of FTDNA, or take a haplogroup panel of YSEQ. Depending on the haplogroup there are considerable differences in the actuality of the panels. You can also skip this step and go for NGS.
Summary, intentions for (2.0):
- Determination of the Y-haplogroup for prehistoric research. (predicted by Y-STR, but also verification by Y-SNP)
- Determination and evaluation of paternal kinship between two men by Y-STR. (Also via database)
3.0 Complete Y-DNA, including haplogroup, terminal-, novel SNPs and Y-STR for genealogy
Since the prices for NGS tests (Next Generation Sequencing) drop, these tests are finding more and more supporters. For those interested in Y-DNA, there are two types of tests. First the Y-based NGS like the Big Y of FTDNA, which only represent the Y-section of the genome (in the case of the YElite of FGC also the mt-section) and then the WGS (Whole Genome Sequencing), which contain the entire genome, including the entire mitochondrial and autosomal DNA.
The advantage of these tests is that not only known SNPs are tested, but almost (apart from so-called “nocalls”) the entire section of Y-DNA with over 23 million base pairs. Almost all SNPs are found that distinguish one from the reference, the human genome. These are about 3000 “positive” SNPs. In special databases, such as Yfull.com, the raw data are analyzed and compared with other testers. Variants, which are shared with others, determine the haplogroup and the terminal SNPs. Variants found for the first time in one subject are called “novel” SNPs or “private” SNPs. These are the biggest advantage of these NGS tests, since one can determine these only in this way, with such a test. These are, so to speak, the Y-SNPs which have mutated over the last centuries and those which have the greatest relevance for genealogy. If another subject uploads his raw data to the same database that is positive for some of these Y-SNPs, a new subgroup is formed in the Y-tree. The “Novel Variants” are also used to estimate the TMRCA of a subgroup. Additionally, several hundred Y-STR (up to 780 for Yfull) can be extracted from NGS tests and their advantages can be used.
The Big Y from FamilyTreeDNA (FTDNA) enjoys great popularity despite its relatively high price, as existing tests, such as the Y37, can be extended without sending in a new sample. In addition, FTDNA has a large database for Big-Y with its own Y-haplotree and some small tools for interpreting the results. Unfortunately, this test can only be purchased in combination with the Y111 and not separately. The YElite from FGC offers good coverage for Y-DNA and also includes the mt-DNA.
WGS are offered by different companies. Besides the Y-DNA, these include the entire mitochondrial and autosomal DNA. Companies like Full Genome Corporation (FGC) and YSEQ are specialized in the genealogical use of WGS raw data and prepare them accordingly, so that I would like to recommend these companies for people who are not able to do these steps themselves. If you trust yourself, you can also choose any other company that offers WGS, provided you get the raw data in FASTQ or BAM formats.
Regardless of which provider you did your NGS with, I recommend the analysis at Yfull.com. This offers so many advantages that it requires several articles to show them. Here is a current blog post with the most important advantages of Linda Jonas: Advantages of submitting to Yfull
A comparison of the different NGS with regard to coverage and their significance for age estimation can be found here: ydna-warehouse.org/statistics
Summary, intentions for (3.0):
- All positive SNPs will be determined, including the “private” variants which have not been tested positive in any other subjects before.
- Extraction of several hundred Y-STR.
- Determination and evaluation of paternal kinship between two men by Y-SNP, Y-STR and private SNPs.
- Active participation in “growing” the Y-trees, such as forming new branches.
- Determination of the Y-haplogroup and subgroup for prehistoric research, but also migration movements of the last centuries.
- Raw data for submitting to Yfull.com.
- Automatic update of the terminal SNP by adding new subjects to the databases.
- Private variants and young terminal SNP to verify paternal relationships, as described in point 4.0.
4.0 Verify paternal relationship, using known terminal SNPs and “Novel” SNPs from NGS
If you wanted to compare the Y-DNA of two men back in time, you used Y-STR tests with as many markers as possible. If an NGS is available, a new method can be used to determine the relationship between two men using “young” SNPs. However, this variant only works if the number of private SNPs or terminal SNPs is not too high, as the costs for this increase with the number of SNPs to be tested. This is made possible by the company YSEQ.net and the possibility to actually test any SNP that can be tested with the Sanger method for little money.
First you have to check if the variants you want to test are already available. With „Wish a SNP“ you tell YSEQ which SNPs you wish for order. You only have to consider that these are in the testable areas. You first verify by testing the terminal SNP that there is actually a paternal relationship. In case of relation, all existing private variants are tested with this test person. Of these, some should be positive and others negative. The number of relatives can be used to evaluate the relationship.
Great article! Loved it!
Pingback: Take part at the Y-tree – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
Pingback: Y-haplogroup from atDNA raw data – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
I would like your permission to use the DNA overview diagram (Y-DNA, atDNA, mtDNA) to use on my DNA website page.
Check:
https://www.kloosterman.be/who-am-i/my-dna-origins/
Hello
yes, feel free to use it.
Thank you!!
You can see the result here:
https://www.kloosterman.be/who-am-i/my-dna-origins/
Hello,
This is very interesting, thank you.
I have questions regarding what you write about the creation of new subgroups by Yfull (in Section 3.0):
“Variants found for the first time in one subject are called “novel” SNPs or “private” SNPs. [..] If another subject uploads his raw data to the same database that is positive for some of these Y-SNPs, a new subgroup is formed in the Y-tree. ”
Do you mean that if someone uploads his data to Yfull and has several “novel” SNPs then no new subgroup will be created? and that in this case a new subgroup will only be created when someone else who shares some of these “novel” SNPs will upload his data to Yfull?
You need at last two individuals to form a new subclade, who share same variants, noone in this database shared them, before.
Pingback: Genetics Research Results Review [2014-2021] | LUND-IA-K