1. The hidden potential of atDNA tests, for Y-DNA projects.
Autosomal DNA tests (atDNA) do enjoy growing popularity. These are mostly advertised with the ethnicity estimations and offer databases for the so-called “matching” (From my point of view, the biggest advantage of these tests). On the page The DNA Geek there are some statistics. According to these statistics, almost 30 million atDNA tests were performed or uploaded. Some are represented in several databases, so that the number of men tested, can be roughly estimated at up to 10 million testers. There is a great hidden potential for Y-DNA projects.
In the autosomal DNA tests of 23andme and Living DNA, the Y-haplogroup and the mt-haplogroup are displayed directly. Many don’t know that the raw data, of all male atDNA testers contain information (Y-SNPs) for the Y-haplogroup and can be extracted with tools like the Morley Predictor. (Only FTDNA removes these Y-SNPs from the raw data).
Irrespective of which company the atDNA test was done with, it makes sense to have a closer look at the results, because there is much more information in it than some people think. If you get an “old” haplogroup displayed, even though younger branches are tested, it can mean that you are sitting on a rare branch.
Not interested in the Y-haplogroup?
You may not be interested in the Y-haplogroup yourself. Nevertheless, I would like to ask you to briefly deal with this topic, extract your Y-haplogroup and communicate this. So, you could contribute a very valuable piece of the puzzle to someone who is intensively involved in a Y-DNA project.
For example, one of my open questions: “Is there another Greek with the Y-haplogroup I-L38?
My Y-haplogroup is I-L38, and my ancestor in the pure paternal line was from Greece. However, I-L38 comes from northern Europe and until recently, I didn’t know of any other Greek people who had been tested with this Y-haplogroup. However, the fact that I do not know anyone does not mean that there is no other tester. It is quite possible that “the wanted man” did an atDNA test but doesn’t know what to do with his result. That is also the reason why I created this blog. So that he can find and contact me, so that we can exchange ourselves about this Y-haplogroup.
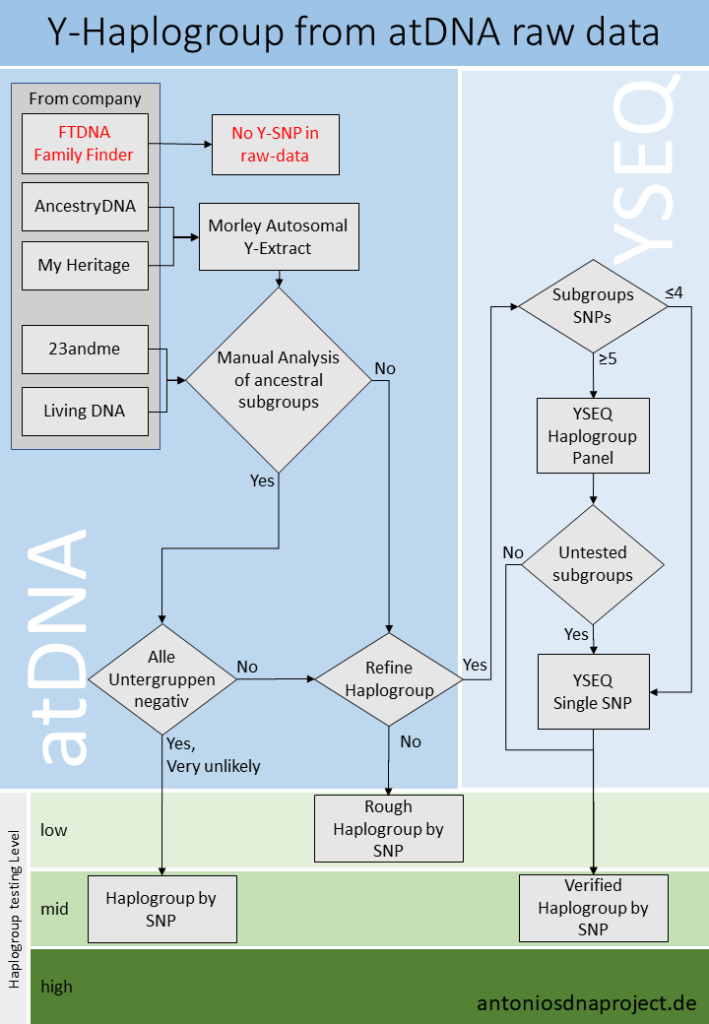
Fig. 1 is a cutting of the overview Y-DNA, Haplogroup and Genealogy
2. What is a Y-Haplogroup?
The Y-DNA is handed over from the father to the sons, only. A Y-haplogroup is a group of men who are all descended from one man, in a purely paternal lineage. The TMRCA (Time to the Most Recent Common Ancestor) in “years before present” is used to date the youngest common ancestor, known as MRCA (Most Recent Common Ancestor). With a simple determination of the Y-haplogroup, the TMRCA can be approximately 5000 years before Present. If you want to know the haplogroup “more precisely”, you can refine it with additional tests on subgroups and possibly come up with an MRCA that lived about 1000 years ago.
In the past, the so-called “long form” was often used for naming Y-haplogroups, which begins with upper case letters and then consists of alternating numbers and lower-case letters. In the meantime, a SNP (Single Nucleotide Polymorphism) for which all group members have been tested positive, is used. My own Y-haplogroup is known as I2a1b2a (long form, valid for 2019) or I-L38 (SNP). Each character of the long form represents an SNP.
| Long Form | I | 2 | a | 1 | b | 2 | a | 2 |
| SNP | M170 | M438 | CTS2257 | L460 | M436 | Y10705 | L38 | S2606 |
| TMRCA (YFull) | 27500 | 21500 | n/a | 20900 | 17100 | 11800 | 4400 | 4100 |
3. Extract the Y-haplogroup from the raw data of an atDNA-Test.
The Y-haplogroup is determined from the Y DNA using SNPs. A higher number of tested SNPs can theoretically lead to a more accurate determination of the Y-haplogroup. In practice, however, it depends on the haplogroup. (In the case of I-L38, Living DNA does not have an additional subgroup compared to 23andme v5, despite almost tenfold SNPs.) One should also know that there are different versions within the companies due to “chip changes”, also related to the number of Y-SNPs. My Heritage changed the chip for its test in spring 2019. The results of the test version 1 (until 03.2019) are to be taken with caution, since only 482 SNPs were tested here, but version 2 is comparable to the test of 23andme v5.
| Test | FTDNA | My Heritage v1 (Bis ca. 03.2019) | My Heritage v2 (ab ca. 04.2019) | Ancestry | 23andme v3 | 23andme v5 | Living DNA v1 | Living DNA v2 |
| Y-SNPs | 0 | 482 | 3524 | 1729 | 1766 | 3733 | 22500 | 34216 |
For the following points I take as an example the raw data from my own tests, Ancestry and 23andme v5.
3.1 Download the raw data and save it to your hard disk.
3.2 Preprocessing, stage 1 of 2
- On the Morley Predictor page, check the box “I consent to the processing and collection of my data, as described in the privacy policy”.
- Select the file.
- Copy the contents of the box “Extracted Y-DNA data” into a spreadsheet or text file.
- Make sure that the box “Errors encountered during processing”, shows “no errors to report”.
- Continue with >>Submit<<.
3.3 Preprocessing stage 2 of 2
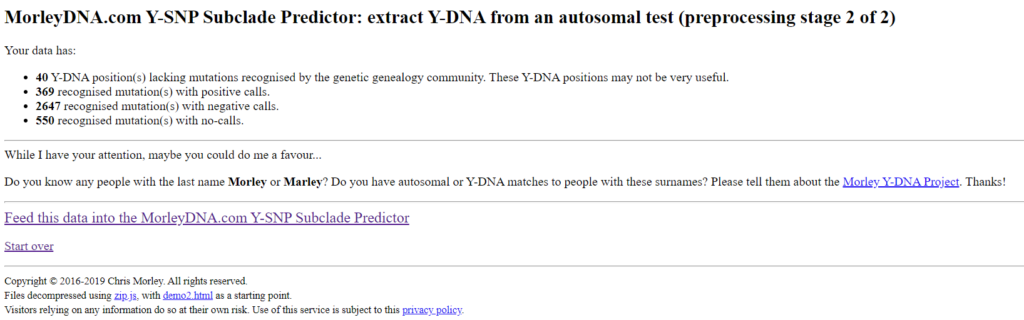
This page contains a list of the processed SNPs and can be used for quality control. If more no-calls are displayed than positive and negative SNPs, there must be an error. For My Heritage Version 2 tests, performed after April 2019, an increased number of „unrecognised position(s). Are you using data from a source other than AncestryDNA, 23andMe or MyHeritage?” is displayed. This is because the Morley Predictor has not yet been optimized for this test. The result is still usable. The following two figures show the values for my tests of Ancestry and 23andme v5.
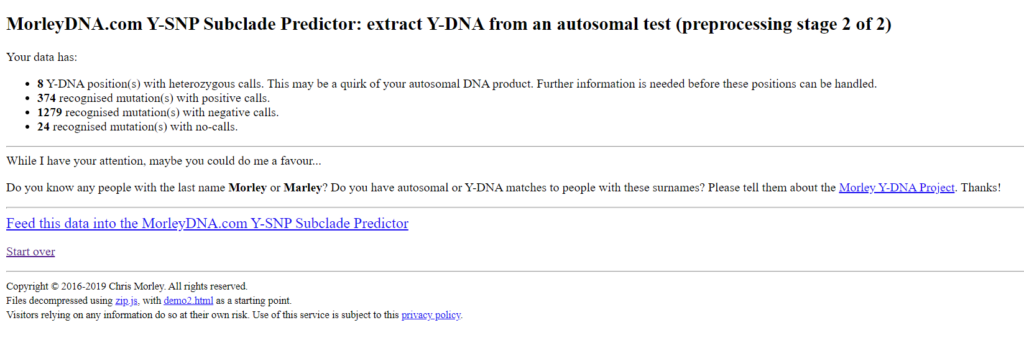
The tests differ in the number of positive, negative and no-calls. It is interesting to note that the 23andme v5 test found fewer positive SNPs, although twice as many SNPs are tested compared to Ancestry. But at 23andme v5 more SNPs are tested for which I am negative, which can also be an advantage.
- Continue with >>Feed this data into the MorleyDNA.com Y-SNP Subclade Predictor“<<.
3.4 List of SNPs
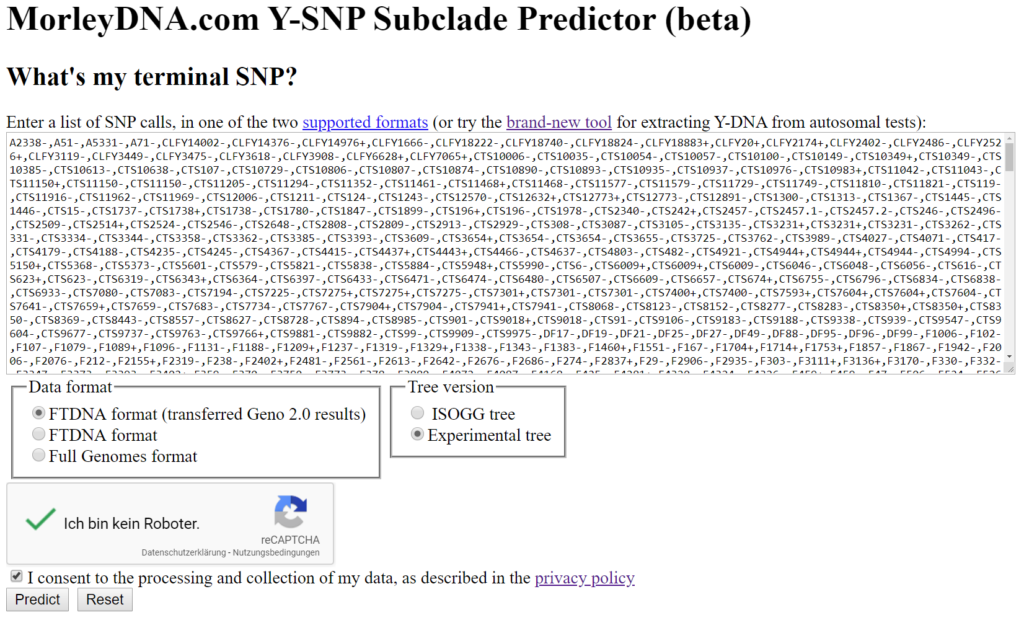
- Copy the list of tested SNPs (contents of the box “Enter a list of SNP calls, ….”) into a spreadsheet or text file.
- Leave the default setting.
- Check the box “I consent to the processing and collection of my data, as described in the privacy policy”.
- Check box, in front of the reCaptcha, unless you are a robot. 😉
- Continue with >>Predict<<.
3.5 The result of the Morley predictor
On the left side you can see suggestions under “Suggested terminal clade”. Usually the first one marked with “most likely” is the right one, but you should look at them all in turn. But really interesting is the right side, if you scroll the way down. (Important! The important information is at the right bottom!) There you can see in a picture, which “younger” SNPs were tested positive and which negative. Fig. 5 shows the test of Ancestry.
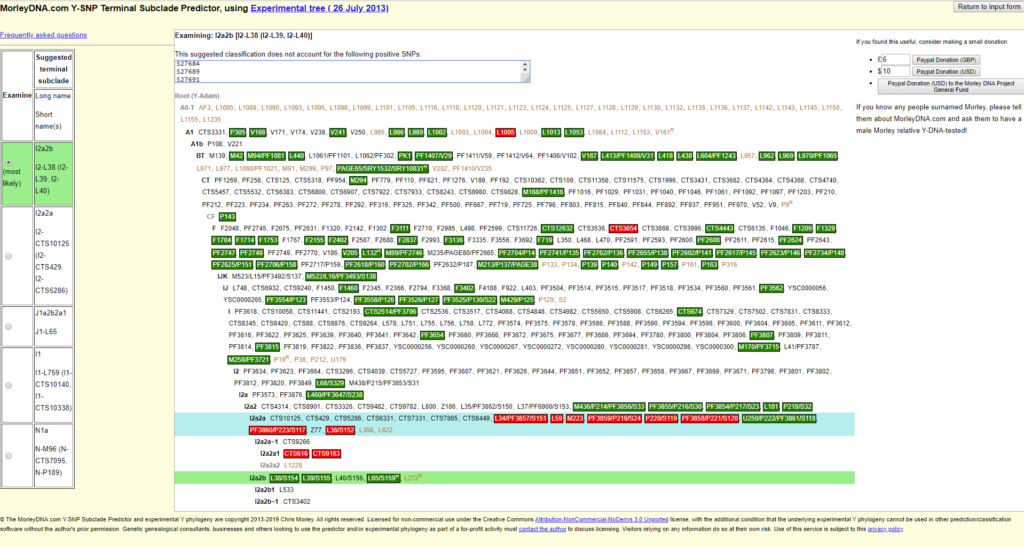
- Behind the name (long form) of the Y-haplogroups in bold letters, are the SNPs belonging to the respective phylogenetic block.
- The positive SNPs are green, the negative ones red, the no-calls and untested SNPs without boxes.
- My determined haplogroup is I2a2b. Under this long form this haplogroup (today I2a1b2a) was known in 2013, when Morley created this subclade Predictor (See: ISOGG Y-DNA Haplogroup Tree 2013). If you use the Long Form, you should write the year in which it was valid. I2a1b2a (2019) from ISOGG Y-DNA Haplogroup Tree 2019, is the same as I2a2b (2013). The short form for this haplogroup is I-L38.
- Check the list above the graph “The suggested classification does not account for the following positive SNPs: ” for positive tested, known subgroups. Here are all SNPs that were tested positive and were not included in the graph.
- Alternatively, the list of tested SNPs, which was saved at point 3.4, can be checked with a Y-tree, e.g. YFull.com, to see if there is any information on subgroups not yet displayed.
I2a2b (I-L38) was identified as the Y-haplogroup (this article uses the Long Form from 2013). Please note the following in this context:
- All tested SNPs behind I2a2b must be positive (green).
- All tested SNPs behind I2a2, I2a, I2 etc. must be positive (green). These are the ancestors of I2a2b and I2a2a (I-M223).
- All tested SNPs behind I2a2a must be negative (red). This is the “brother” of I2a2b and the second phylogenetic child of I2a2.
- SNPs that break these rules must be looked at separately. For example, U250/P222/PF3861/S118 is a SNP which is no longer used (see https://yfull.com/search-snp-in-tree/), as it cannot be placed accurately in the tree. The other two red SNPs L1005 and CTS3654 were tested positive in my NGS test (Next Generation Sequencing) and are not shown correctly here.
It does not always have to be about errors. If green and red fields are mixed in the block of the detected haplogroup (in my case I2a2b), this can in rare cases mean that you belong to a very rare (not yet detected) branch (e.g. of I2a2b). More under 4.2.
The above points are also valid for the test of 23andme v5, see Fig. 6. If you compare the two figures, you can see that other SNPs were tested. At the bottom you can see that the subgroup L533 was tested negative.
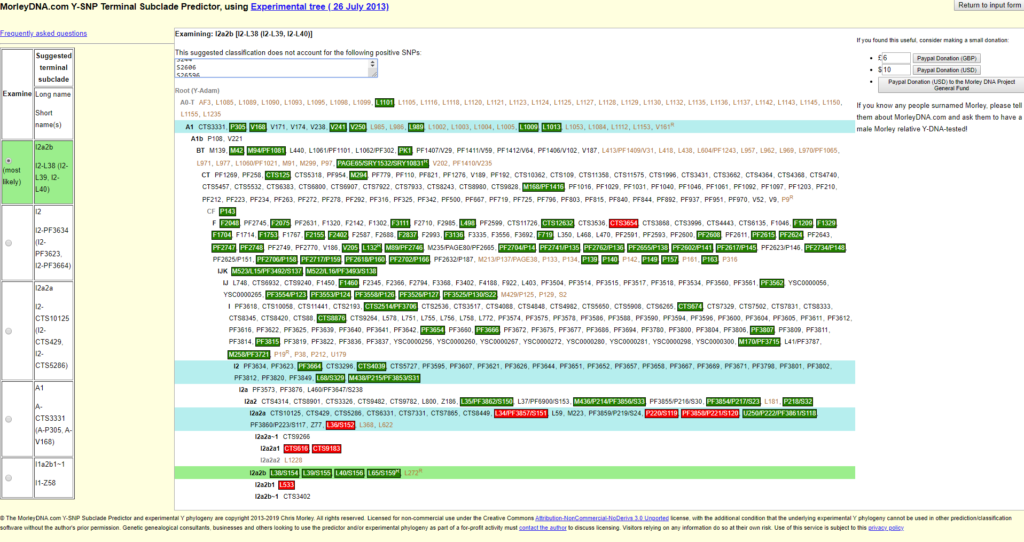
This is not the only subgroup tested by 23andme v5. My Y-haplogroup displayed on 23andme’s website is I-L38>S2606. This SNP (S2606) is not shown in the graphic of the Morley Predictor from 2013, because it was not known at that time. The SNP S2606 appears in the list above the graphic “The suggested classification does not account for the following positive SNPs:” and can also be found in the list of 3.4 as positive SNP.
In various forums, people ask the question “How deep does this Y-DNA test go?”. In my example, 23andme v5 tests to I-L38>S2606, with a TMRCA of 4100 ybp. (years before present). Age estimation from YFull (yfull.com/tree/I-L38/).
4. Additional information through “negative subgroups”.
The atDNA tests are used to test many Y-SNPs. Some of them are tested positive, others negative. With the positive ones, you find the path to your Y-Haplogroup-SNP. You can overlook how important it is to know the SNPs that were tested negative in a test. Especially if the TMRCA for the determined Y-haplogroup is very old, there can be very interesting reasons. You might belong to an old, rare Y-haplogroup.
For the following points only my test of 23andme v5 is considered. You can use the files saved in point 3.4 and point 3.2 for further analysis to query SNPs not shown in the graph.
- The list of 3.4 is easier to use. It contains all tested SNPs with a plus for positive and a minus for negative. If no SNP was detected for the tested hg19 position, the hg19 position is specified.
- The list of 3.2 contains the position hg19 and the value of the test for this position. (derived, is the changed value if positive). On the site genetichomeland.com you can see the position hg19 for the SNPs.
Example A in the table stands for “Antonios” and shows my values as determined in point 3. The examples F1 to F4 are fictitious. We imagine that these SNPs would be tested in a test like this and interpret the results.
| Long Form and SNPs | Pos. hg19 | Ref. | derived | A | F1 | F2 | F3 | F4 |
| I2a2 (I-M436) | ||||||||
| L35/PF3862/S150 | 22725379 | C | A | + | + | + | + | + |
| M436/P214/PF3855/S33 | 18747493 | G | C | + | + | + | + | + |
| PF3854/P217/S23 | 7628484 | C | T | + | + | + | + | + |
| P218/S32 | 17493630 | T | G | + | + | + | + | + |
| I2a2a (I-M223) | ||||||||
| L34/PF3857/S151 | 7716262 | A | C | – | – | – | – | – |
| P220/S119 | 24475669 | G | T | – | – | – | – | – |
| PF3858/P221/S120 | 8353707 | C | A | – | – | – | – | – |
| I2a2b (I-L38) | ||||||||
| L38/S154 | 15668070 | A | G | + | + | + | – | + |
| L39/S155 | 16199051 | T | C | + | + | – | – | + |
| L40/S156 | 16202267 | T | C | + | + | – | – | + |
| L65/S159.2 | 16626617 | A | G | + | + | + | – | + |
| I2a2b1 (I-L533) | ||||||||
| L533 | 2887198 | G | C | – | – | – | – | + |
| I2a2b2 (I-S2606) | ||||||||
| S2606 | 22527402 | C | A | + | – | – | – | – |
This table includes only SNPs tested by 23andme v5. The results of both tests could also be combined here. In the second column are the hg19 positions, in the fourth column the value for this position as stored in the raw data. Fig. 7 shows the possible positions on the Y-tree of YFull, depending on the (fictitious) determined values.
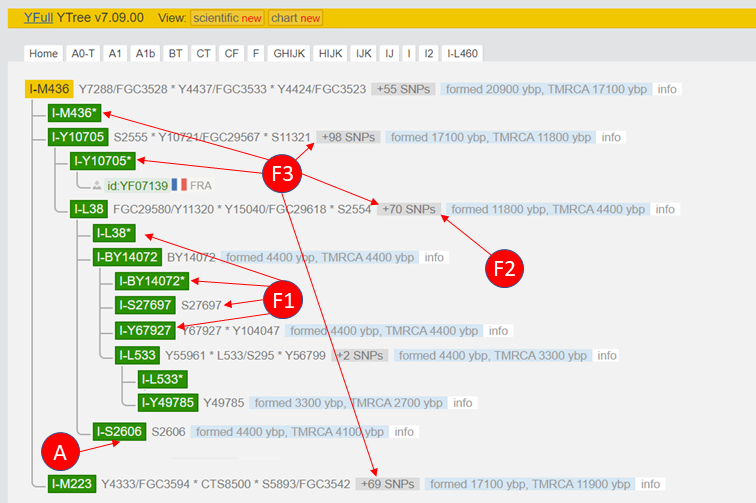
4.1 Example F1: Subgroups of I-L38 are negative.
When testing 23andme v5, two SNPs are tested for the subgroups of I-L38. If either S2606 or L533 is positive, the result is clear (see example 0 and F4). Note that L533 is no longer directly below I-L38, but a subgroup, the subgroup BY14072.
If both SNPs are negative, there are three possibilities:
- The tester is I-L38*, an undiscovered subgroup of I-L38. (unlikely, but possible)
- The tester is I-BY14072*, an undiscovered subset of I-BY14072.
- The tester belongs to one of the subgroups of I-BY14072, S27697 or Y67927.
(The Ancestry test does not test SNPs for subgroups of I-L38.)
4.2 Example F2: Some of the SNPs of I-L38 (I2a2b) are negative.
This case is also very unlikely, but not impossible and would be a real stroke of luck for our I-L38 haplogroup project.
The Y haplogroup I-L38 is defined by 73 SNPs. Four of them are tested in the test of 23andme v5. To belong to the haplogroup I-L38, one must be positive for all SNPs of this phylogenetic block. If you are negative for one or more of these SNPs, this means that you divide the phylogenetic block of I-L38 into two blocks. This happened before when YF07139 (Fig. 7) was added to the tree. Previously all SNPs of blocks I-Y10705 and I-L38 belonged to block I-L38. See blog post NGS – Take part at the Y-tree.
If such a case occurs, you should show these results to someone who can evaluate them more accurately. You can search the Y-haplogroup projects at FTDNA and contact an admin of the group or join a group for your Y-haplogroup at Facebook.
In the case of I-L38, this would be the I-L38 haplogroup project and the Facebook group I-L38. (Of course, it would also be possible to contact me via this blog).
4.3 Example F3: All SNPs of I-L38 (I2a2b) and I-M223 (I2a2a) are negative.
This case is, like example 3, very unlikely and therefore very interesting for the Y-Haplogroup projects I-L38 or I-M223. The possibilities in this case would be:
- Splitting the I-L38 block of SNPs, in the early stage
- Splitting Block, I-Y10705
- Form a subgroup with YF07139.
- New undiscovered subgroup under I-M436
- Division of block I-M223, early stage
If such a case occurs, you should show these results to someone who can evaluate them more accurately. You can search the Y-haplogroup projects at FTDNA and contact an admin of the group or join a group for your Y-haplogroup at Facebook.
In the case of I-L38, this would be the I-L38 haplogroup project and the Facebook group I-L38. (Of course, it would also be possible to contact me via this blog).
5. Possibilities for extending and deepening the results
The Y haplogroup information you get from an atDNA should be sufficient for many. If you want to deal with the topic more intensively, you can take the results as a basis and extend them with different products.
5.1 YSEQ Haplogroup Panel and Individual SNPs
The company YSEQ from Berlin offers a very flexible product range for this purpose. Here you can find so-called “Haplogroup Panels”, but also single SNPs, which can be ordered for testing (see fig. 1, right).
Haplogroup panels are SNP packages with a sophisticated selection of SNPs, of a haplogroup. The individual SNPs are tested sequentially until you reach your most downstream position. This is the case if all known subgroups are tested negatively. The suitable one for I-L38 would be the I2 Superclade Panel (detail Fig. 8).
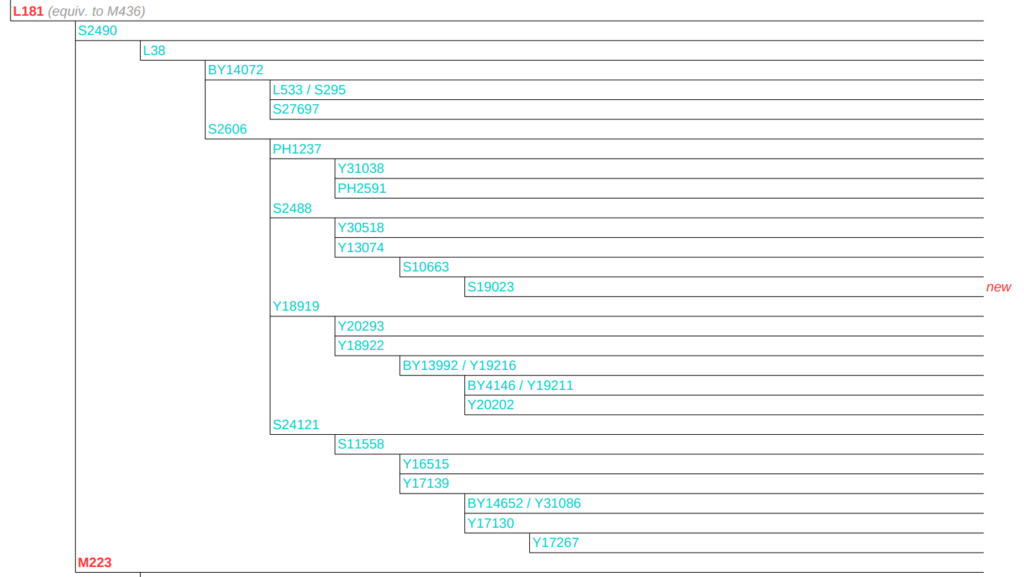
The panels only make sense if there are enough subgroups that have not yet been tested. The I2 Superclade Panel costs $99. A single SNP costs $18. A Superclade Panel only pays for itself with 6 SNPs or more. (Haplogroup panels for $88 pay off from 5 SNPs). For this we take the examples from point 4.
5.1.1 Example A > Haplogroup Panel
In example A this panel makes sense. There are many SNPs to test. This panel would determine Y31038 as terminal SNP for me. But there are other subgroups of Y31038 which are not tested on this panel. These subgroups can be tested afterwards (if available).
5.1.2 Example F4 > Single SNPs
In the example F4 the panel makes no sense. Here you wouldn’t get any additional information at all. You already know that you belong to L533. The known subgroups of L533 are L533>Y49785 and L533>Y49785>S18432 (see Fig. 7 and Y-DNA Haplotree of FTDNA). These two SNPs cannot yet be ordered from YSEQ. With the product “Wish a SNP” you may wish to add this SNP to the YSEQ catalogue.
5.1.3 Example F1 > Individual SNPs
In the F1 example, the panel makes no sense. The SNPs L533 and S2606 were tested negative. There is a maximum of three SNPs to test. Proceed as follows:
- Test SNP BY14072.
- If BY14072 is negative, then you are L38* (only one SNP is tested).
- If BY14072 is positive, then test all subgroups except L533 (= negative) (S27697 and Y67927).
- If S27697 and Y67927 are both negative, then you are BY14072*.
- If one of them is positive, then this is your terminal phylogenetic position.
If your result is I-L38*, you should show these results to someone who can evaluate it more accurately. You can search the Y-haplogroup projects at FTDNA and contact an admin of the group or join a group for your Y-haplogroup at Facebook.
In the case of I-L38, this would be the I-L38 haplogroup project and the Facebook group I-L38. (Of course, it would also be possible to contact me via this blog).
5.2 NGS (Next Generation Sequencing)
In the examples F2 and F3, you cannot expand with simple means. The only reasonable possibility is to test an NGS and help to discover new branches in the Y-haplotree. How this works can be found in the blog post NGS – Take part at the Y-tree.
6. Benefit, Y-DNA from atDNA raw data
- Determination of the Y-haplogroup for prehistoric and early historical research.
- While these tests do not indicate that two men are related in the paternal line, they do indicate that two men are *not* related in the paternal line, which can often be helpful.
- Due to the large databases and the associated autosomal matches one can also use this well for “fishing”, i.e. finding testers who are relevant for one’s own Y-research and contacting them.
I took a test at Livingdna and only got an overall score of E-V13. In the Morley predictor, I got the result PF1919 and I don’t know what to do next. Can you help me do a deeper analysis? I can send the file to your e-mail address, but I couldn’t find the address on the website.
Please don’t send the file. I won’t open it. Thanks.
I mailed you a processed file for the Morley predictor test. You have a lot of knowledge, maybe you can determine for me a more precise subclade of the haplogroup. I will be grateful for your help.
Can you explain to me the panel purchasing service at Yseq? Do I need to do the swab test again or will I just need to send it to them with my data? I tested again (at Ancestry) can I order the ydna panel now?
Hello Artur,
Ancestry is a company in USA, while YSEQ is a different company in Berlin, Germany. Surely they need a swab from you.
Thanks for the explanation. So I didn’t need to do the Ancestry test since I got the same result from the Livingdna test.
Pingback: Family Finder Y-DNA Haplogroups – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
Someone with Y-dna haplogroup J and someone with Y-dna haplogroup Q share many SNPs but also at the same time contained different SNPs that are unique to J and Q, is that correct?
For instance, here is the Y-SNP calls for Barcin 31 https://genetiker.wordpress.com/y-snp-calls-for-barcin-31/.
According to the link, Barcin 31 is positive for A00-FGC27424/YP3511, A00-M8129, A0-T-YP1873, B-M8633-M182-M150-M6473, G-P287-P15-L1259-L30-CTS1180/PF3259/M3244, IJK-PF3500/M2696, J-PF4513, Q-L274-L472-F1096-M25-L712-L715-YP836, R1b-L278-L389-P297-M269-PF5462 and so on but eventually “The calls show that Barcın 31 belonged to Y haplogroup G2a2b-L30*”.
Is it correct if we assigned this individual as A00, A0-T, basal J or any of the Y-dna haplogroups above because the individual have SNPs belong to other Y-dna haplogroups? If it is not, why?
The J man shares the SNPS with the Q man, which are from A to IJK.
The IJ and J SNPs are From J man only and Q man has none of them and the J man has not the SNPs from K to Q.
You look at ancient DNA. It isn’t easy to place those samples correctly, especially if the quality isn’t good and there are lots of false positive and false negative.
Living men with good WGS test don’t have this problem and you can find the terminal SNP.