I did a DNA test in early 2017. My main concern was to explore a family legend in my paternal line. I wanted to know if it was true, that my ancestor in my paternal line was indeed a man named Gerakaris, who came from Mani (My parents come from Northern Greece, Mani is in Southern Greece). So, from the beginning my focus was on the Y-DNA. At the beginning I approached the topic very naively and thought that a single Y-DNA test could help me, and ordered a so-called Y37, together with a test for atDNA and mtDNA.
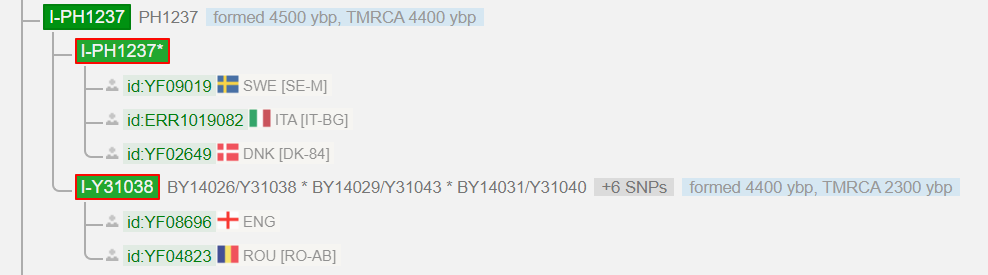
I got 37 Y-STR markers that I couldn’t do much with and a prediction for a Y-haplogroup. This was I-M170, with a TMRCA (Time to Most Recent Common Ancestor) of 27500 ybp. The company FTDNA is known for the fact that the predictions are very conservative. Later, these markers were interpreted by a haplogroup administrator as I-L38 > S2606 > PH1237 (TMRCA = 4400 ybp), a haplogroup found mainly north of the Alps. He also suspected that I could be the first from Greece, to belong to it’s small southeast European branch. This was soon confirmed using a SNP pack. My confirmed haplogroup was now I-L38 > S2606 > PH1237 > BY14026/Y31038 (TMRCA = 2300 ybp). Was that all? Really? How did this Y-haplogroup get from Northern Europe to Greece? The Y-tree of YFull ended at Y31038 at that time, and there were just three men on that branch: Two Englishmen and a man from Romania. I asked myself: “Isn’t there a test that goes on, so that you can add more branches to the trees?
The answer was “Yes, the NGS tests”.
This article deepens the topic NGS. An overview of the possibilities for testing Y-DNA can be found in the article Y-DNA, Haplogroup and Genealogy.
1. NGS – Next Generation Sequencing
There are two types of NGS (Next Generation Sequencing) tests. Such as the Big Y of FTDNA or the YElite of FGC, which specifically reads only the genealogically relevant regions of the Y chromosome (green box, Fig. 2) and the WGS (Whole Genome Sequencing) which, as the name suggests, reads the whole genome, including all mitochondrial and autosomal DNA.
1.1 Targeted NGS
The Big Y from FamilyTreeDNA (FTDNA) is very popular despite its relatively high price, as existing tests, such as the Y37, can be expanded without sending in a new sample. In addition, FTDNA has a large database, its own Y-Haplotree and some small tools for interpreting the results.
Full Genome Corporation’s (FGC) YElite also includes mt-DNA and data analysis.

1.2 WGS
WGS are offered by different companies. In addition to the Y-DNA, these contain the entire mitochondrial and autosomal DNA (mtDNA & atDNA). Companies like Full Genome Corporation (FGC) and YSEQ have specialized in the genealogical use of WGS raw data, and prepare them accordingly, so that I would like to recommend these companies for people who are not able to do these steps themselves. If you are confident in yourself or know someone who can help you, you can also choose any other company that offers WGS, provided you get the raw data in FASTQ or BAM formats.
A comparison of the different NGSs in terms of coverage and their significance for age determination can be found here: ydna-warehouse.org/statistics
1.3 What do you get?
In these tests, not only a small selection of the Y chromosome is tested, but also the entire genealogically relevant area with over 23 million base pairs, depending on the provider and test. The sequence obtained is aligned with the human genome reference and the values of the tester are compared with the human genome reference. The current version is HG38, but occasionally the older version HG19 is still used. For each of these base pairs there is a number for the position (these differ from HG19 to HG38) and the values determined for this position in the form of letters (A, T, G and C). If such a position differs from the reference (“Sample allele” different to “Reference allele”) , it is called SNP (Single Nucleotide Polymorphism). Fig. 3 shows the SNP L38, where there was a change from A to G at position 13556190 (HG38).
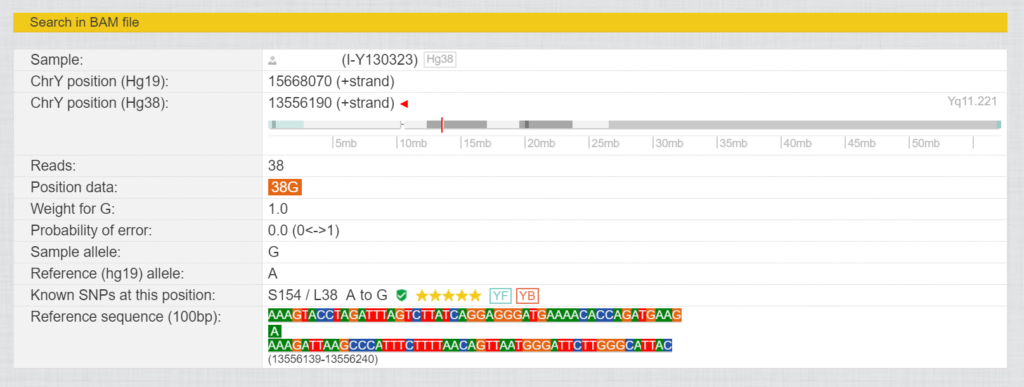
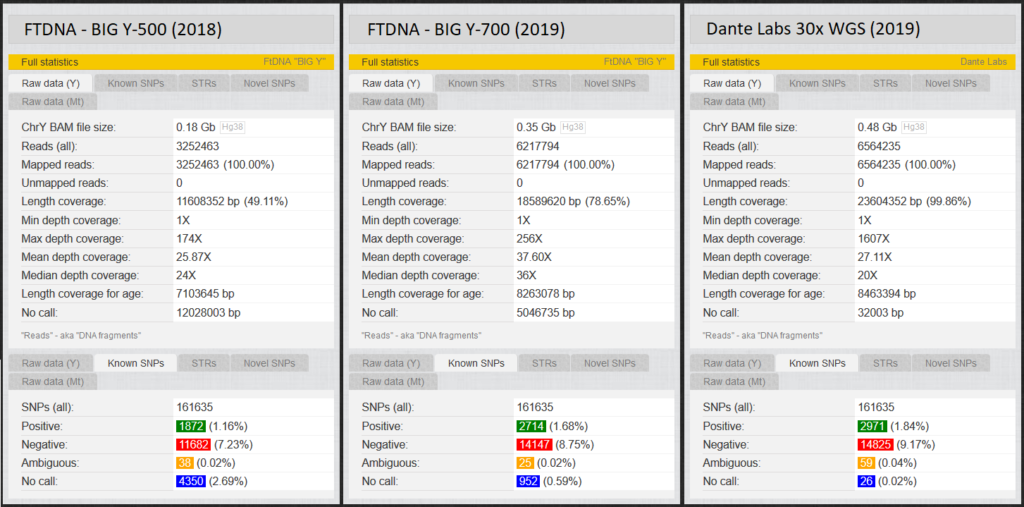
Fig. 4 shows the “Length Coverage” that, depending on the test, different numbers of these approximately 23 million base pairs are tested. “Depth coverage”, on the other hand, indicates how often the positions were read. At YFull, just over 160,000 of these base pairs are known as SNPs. Each of us men should have about 2,300 positive Y-SNPs. (YFull counts some SNPs multiple times if they have different names. Therefore 3000 > 2300). Positions that were not read during a test are called “no calls”. Accordingly, it is not known whether a positive or a negative value exists for this variant, so that a test with the largest possible coverage (length coverage) and few no calls is aimed at. We will later look at an example of the effects of these no calls.
In addition, several hundred STRs (Short Tandem Repeats) can be extracted from NGS tests and their advantages can be exploited. With YFull this is up to 780.
1.4 Raw data:
1.4.1 FASTQ:
This file contains all the raw data and has not yet been aligned with a reference. If you don’t have the “right” BAM, you can create it yourself with the help of this file. The programs for this run under the Linux operating system and the creation takes several days. Alternatively, you can do this action at Sequencing.com or for free at usegalaxy.eu. Please note that this is only reasonable with a fast internet connection, because the FASTQ can be over 60 GB in size.
1.4.2 BAM:
This is the file needed to participate in the trees. It contains the data aligned with the reference. If you did your NGS with a company, like FTDNA, FGC or YSEQ, which are specialized in the genealogical application of the NGS, you will get a link, which you simply can link to when ordering at YFull. Here you can ignore the rest of the paragraph.
For all, who made their
WGS, with another company, the following is of importance:
The current version of the reference is HG38, but occasionally the older
version HG19 is still used. The preferred version for the transfer to YFull is
the HG38, but you can also use the HG19 if you don’t have the HG38 available,
or if you’re not able to create it yourself from the FASTQ. In the same
directory as the BAM, there should be a file with the extension BAI. This is
the index file.
A BAM from a WGS contains, besides the Y-DNA, the complete mtDNA and atDNA and is up to 100 GB in size. To participate in YFull only a small part of it is needed, the sections of the Y-DNA and the mt-DNA. If you have experience with Linux, you can create a file with the program Samtools, which only contains this portion. Windows users can access a tool called WGS Extract (link to download). This allows you to create this file intuitively and make it available later via your own cloud. With WGS Extract you can also create a FASTA file with the mtDNA or extract the atDNA and save it in different formats.
A description of the program WGS Extract can be found here in this blog: http://www.beholdgenealogy.com/blog/?p=3018
1.4.3 VCF:
The VCF format contains only part of the raw data. It is difficult to say exactly what is contained in this file, because it depends on the company. The most beautiful saying I had read was: “Don’t trust any VCF that you didn’t manipulate yourself”.
After people have to pay for the creation of a BAM at FTDNA, since November 1st, YFull decided to also allow the vcf file of FTDNA to participate in the tree in the future.
If, after making a WGS, the BAM is not yet available, some Y information can be read from this file. With the DNA Kit Studio you can extract the Y information from the VCF and analyse it with the Morley Predictor.
1.5 Advantages of NGS
- All positive SNPs will be determined, including the “private” variants which have not been tested positive in any other subjects before.
- Extraction of several hundred Y-STR.
- Determination and evaluation of paternal kinship between two men by Y-SNP, Y-STR and private SNPs.
- Active participation in “growing” the Y-trees, such as forming new branches.
- Determination of the Y-haplogroup and subgroup for prehistoric research, but also migration movements of the last centuries.
- Raw data for submitting to Yfull.com.
- Automatic update of the terminal SNP by adding new subjects to the databases (Y-trees).
- Private variants and young terminal SNP to verify paternal relationships, as described in point 4.0 of the article Y-DNA, Haplogroup and Genealogy.
2. The Y-tree
2.1 The Y-Haplogroup Trees:
- If you have made your NGS with FTDNA, you will be placed in the in-house Y-haplotree after completion of the test.
- Y-Haplogroup R1b testers can upload their raw data to Alex Williamson’s The Big Tree.
- At the Y-tree of https://yfull.com/ every tester, independent of the Y-haplogroup and the provider of the test, can submit his raw data. This tree is my favorite.
- The ISOGG tree is updated regularly.
2.2 A line on the Y-tree from Adam to Antonios
What does such a line look like on the tree? The 2300 positive SNPs obtained from an NGS can be imagined as a chain of changes between the so-called reference chromosome (“Adam”) and the tester. One can roughly assume that one SNP has changed every 80 to 100 years. The figure shows the first 1,949 SNPs, which were formed from “Y-Adam” to the MRCA (Most Recent Common Ancestor) of I-M170, in the blocks AO-T > A1 > A1b > BT > CT > CF > F > GHIJK > HIJK > IJ > I-M170. These are very old SNPs, which were created many thousands of years ago. All men belonging to the Y-haplogroup are positive for these nearly 2000 SNPs.
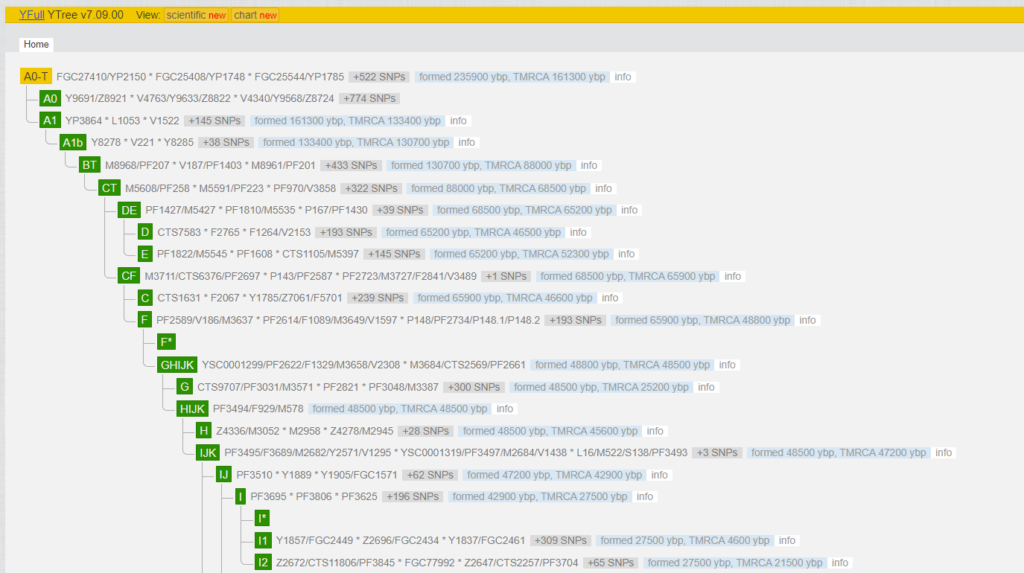
The cut-out is for I-M170. The whole tree can be found under YFull in different views (classic, scientific and chart). It is worth taking a closer look. The remaining 348 SNPs are between the MRCA of I-M170 and the MRCA of my subgroup I-Y130323. I2-M438 > CTS2257 > L460 > M436 > Y10705 > L38 > S2606 > Y13067 > Y13076 > PH1237 > Y31038 > BY25359 > Y125026 > Y128714 > Y130323. Here you can see the rough order of the SNPs, because the order within these blocks is not (yet) known.

Up to SNP Y31038 (BY14026) I removed all kits from the view to get a better image of this line. This is exactly the SNP that was originally determined by the SNP pack. What you see below would have remained hidden without NGS. (Note the comparison to April 2017, Fig. 1.) This is the interesting part for me – The “latest” 26 variants (=SNPs), which have changed in the last two thousand years. These are divided into the four blocks BY25359 > Y125026 > Y128714 > Y130323 and are used for further investigations. (The kits under Y130323 have no “private” variants worth mentioning.)
My line is only one of many. A tree becomes the whole thing only if several lines are linked with each other. I hope that in the next years the prices for the WGS will fall and more men will participate in Y-tree projects and that the trees will grow even “more beautiful”. I also hope that the people who made a WGS for other reasons will participate with their raw data in the project of YFull.com and support the phylogenetic research.
2.3 What happens if you participate in a tree?
You take part in a Y-tree by, for example, performing the NGS of FTDNA and thus accessing its haplotree, or by transmitting the raw data of an NGS (also WGS), independent of the vendor, to YFull or “The Big Tree”. In the databases, your results are compared with the existing participants, and your line, from the tree trunk to your branch, is integrated into the tree.
Most of your variants are already known SNPs in the database. Based on these, you will be assigned a temporary place on the Y-tree. In addition, you have some “new” SNPs, also called “private” SNPs, which have not been found by any other tester before. After the more detailed analysis you will take your exact position in the tree. There are different situations that can occur. These I want to explain to you with the help of fig. 6. (Points 2.3.1 to 2.3.3)
2.3.1 Sharing all SNPs:
YF63639 and YF65688 (YF15460 before upgrade) are positive for all variants for which YF11483 is also positive. The participants have the same Y-DNA for their SNPs. None of the participants has private SNPs left. Therefore, the TMRCA for this group is estimated at 50 “years before present”. (The real TMRCA is about 150 to 200 years.)
Although the SNPs are the same, there may be some differences in the STRs. If this is the case, these can be used for further grouping below the MRCA.
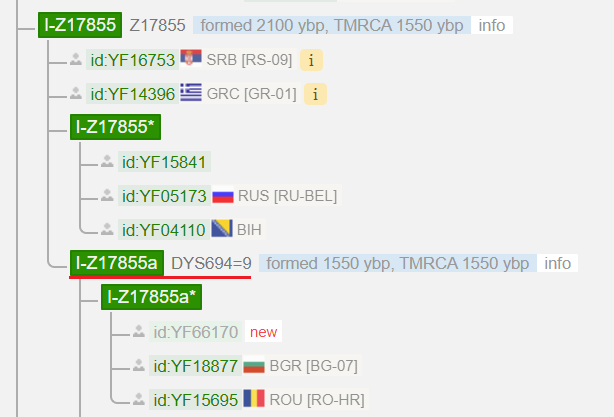
2.3.2 Create a generic subgroup:
YF19586 Is positive for all SNPs within the block of Y125026 (Applies to all Y125026*). He does not share any “private” variants, and is, so to speak, the only participant of a future subgroup. These are marked with an asterisk “*”.
A possible reason why no variants for subgroups were found may be that they were not tested due to a poor coverage of the tests (e.g. Big Y-500) and thus increased no calls. This situation can be improved by upgrading to a more advanced test, such as a WGS or a Big Y-700. However, there is no guarantee that new private SNPs will be found. In the case of 2.3.1 I could not find any private variants despite the upgrades.
(A look at the Y-STRs suggests that Y125026 participants could be divided into two groups based on a specific STR (DYS448, 4 stars). See 2.3.6)
2.3.3 Sharing of some private variants:
YF65013 had more luck. HGDP00894 (participant of a scientific study) was already in the database before than Y125026*. For seven formerly private SNPs of HGDP00894, YF65013 is positive. These now determine the new subgroup Y177573.
2.3.4 Detaching individual SNPs from a block:
When YF63287 was added, the subgroup Y125026 did not yet exist. At that time this was still known as BY25359. All participants of this subgroup were positive for the variants, which are now under Y125026, as well as for BY25359.
YF63287 is only positive for BY25359, but negative for the SNPs below Y125026. As a result, the block BY25359 became Y125026 by removing the SNP BY25359. It was just a coincidence that the oldest SNP was chosen to name the block.

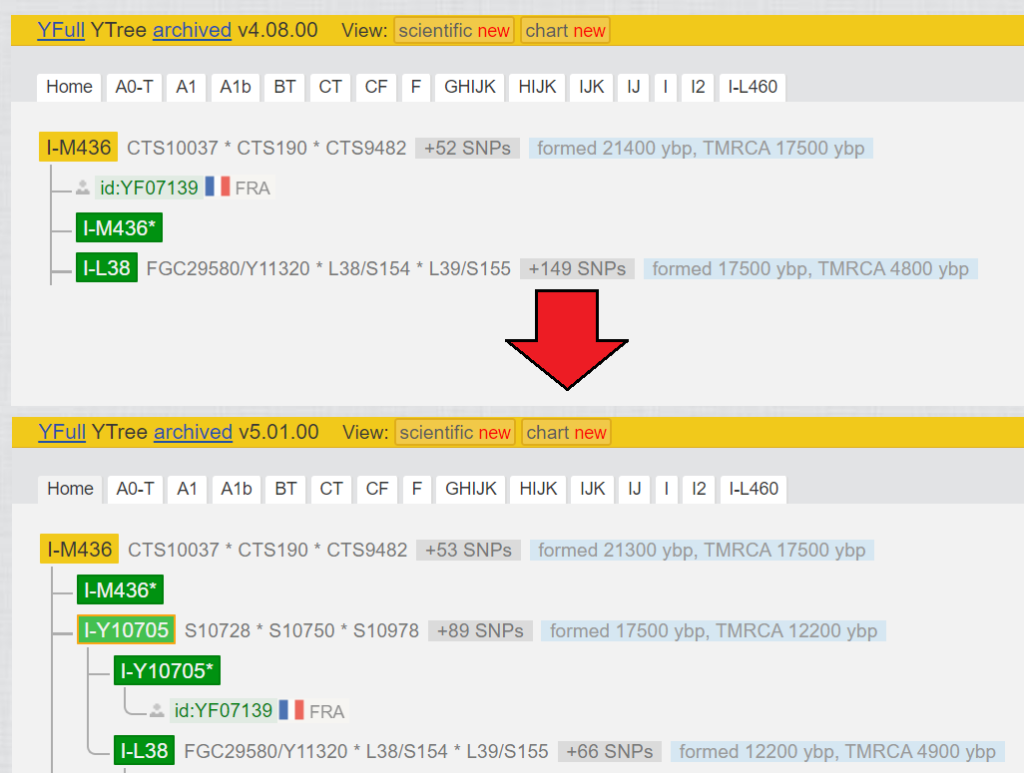
2.3.5 Split one block in two:
Actually, the same as in point 2.3.4, but with drastic effect you can see in Fig. 8. The block of the Y-haplogroup counted 152 variants, with a bottleneck of 12700 years, until YF07139 was added.
- SNPs for which YF07139 is negative were added to the block of I-L38.
- SNPs for which YF07139 is positive were added to the block of I-Y10705.
- in addition, there were a lot of SNPs which were no calls when testing YF07139 (old Big Y-500). These could not be positioned accurately and were evenly distributed among the blocks I-L38 and I-Y10705. These can only be positioned correctly if either YF07139, or someone in the same position I-Y10705* uploads a test with better coverage (Length coverage).
2.3.6 Branches defined by STR
Up to now it was only about branches, which are defined by SNPs. These can also be defined by STRs, which have different mutation rates. If a STR mutates very rarely (low mutation rate), it can also be used to distinguish and define branches. YFull uses STRs with the mutation rate of “five stars” in their tree.
2.4 Naming the new variants and SNP blocks
2.4.1 Designation of the variants:
If new variants are added to the tree, they are named by the company or person who “discovered” them. Depending on the “discoverer”, different prefixes are used here. The most common ones now:
- A = YSEQ.net
- BY = Big Y-500 from FTDNA
- FGC = Full Genomes Corp. (FGC)
- FT = Big Y-700 from FTDNA
- Y = YFull.com
A complete list of prefixes can be found at ISOGG.org.
It can happen that SNPs are “discovered” and named by several companies at the same time. These are often written together with a slash (e.g. Y128714/BY35091).
2.4.2 Labelling SNP blocks
SNP blocks are labelled after a containing SNP.
Again, there are no matching rules and it can happen that different SNPs are used to label blocks on different Y-trees. A good example is Y128714. This block even has three labels:
- YFull = Y128714
- FTDNA = BY35090
- ISOGG = A577

Within the blocks we cannot determine the order of the SNPs. The SNP used for labelling can therefore be the oldest, the youngest, but also one in between. This is only possible if the block is divided by another test.
2.4.3 ISOGG Longform
Some of you know the writing form for Y haplogroups, which starts with an uppercase letter and then alternates between lowercase letters and numbers (Fig. 10). If you take this form, I-L38 currently has I2a1b2a for I2-M438 > CTS2257 > L460 > M436 > Y10705 > L38.
| I | 2 | a | 1 | b | 2 | a |
| M170 | M438 | CTS2257 | L460 | M436 | Y10705 | L38 |
If you use this form of writing, you should absolutely indicate the year, since this is regularly adjusted. When I started 2017, I-L38 was still I2a2b and not long before I2b2. This spelling is very pictorial, but I don’t like to use it because of confusion.
2.5 Why are the branches on Y-trees different, from company to company?
If you compare the Y-trees of YFull and FTDNA, you will notice that branches may be missing. This is because the databases can only process this information if they get it. The reasons are:
- The test was done on FTDNA and not added to YFull or “The Big Tree”. This could be avoided if everyone submits their Big Y to YFull. You can ask the testers to submit their raw data to Yfull. I do this occasionally for the subgroups of BY25359.
- If the information is missing from FTDNA’s haplotree, it is because the test was done by another company. FTDNA does not offer an upload option.
3. How to support the Y-tree projects
Since my first Y-DNA test I have been fascinated by the Y trees. Especially the YFull tree and the Y haplogroups I-L38 and I-Y3120. That’s why I support these projects and have already contributed some NGS. But there are also some other ways to support haplogroup projects.
- Participation with existing Big Y raw data on all Y trees:
I would like to encourage everyone who made a Big Y, to transfer the raw data to the other Y-trees as well. These are the trees The Big Tree by Alex Williamson, for carriers of the haplogroup R1b, and independently of the haplogroup the tree of YFull.com, who run a mt-tree, also. - Participation with existing WGS raw data on all Y-trees:
If you made a WGS, because of the health information, you will most likely not be interested in this topic. Irrespective of whether you were interested in it or not, I would like to ask you to participate in the above-mentioned trees (YFull and if R1b also The Big tree). You already have the raw data. The reason is simple. Some testers interested in genealogy have been waiting for years for participants who can contribute small but important information to their research. With your help new branches could be formed or SNP blocks could be splitted.
In my case, I am waiting for testers to provide information, so that my last 26 variants can be sorted. I am also interested in the haplogroup distributions of Greece. I am especially interested in the Y haplogroups under I-M170 of the Greeks. - Donations to haplogroup projects:
Volunteer haplogroup administrators, who support the projects in their spare time, are engaged in haplogroup projects. In these projects at FTDNA there is the possibility to donate via Group General Fund Contribution. Numerous projects can be supported here (link is for I-L38). Mostly this money is used to extend the test of a project participant whose STR markers point to a previously undiscovered old subgroup.

Pingback: Y-haplogroup from atDNA raw data – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
Pingback: Verify relationship in paternal line, with known terminal SNP and private „novel“ SNPs from NGS – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
Pingback: Y-Απλοομάδα από ακατέργαστα δεδομένα atDNA (αυτοσωματικού DNA) – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
Pingback: YFull – First Steps – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
Pingback: I-L38 – FigUre Tree – 2020-v8.07 – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
Pingback: Y-Haplogroups and migration, using the example of I-L38>BY14026 – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
Pingback: Y-απλοομάδες και μετεγκατάσταση, χρησιμοποιώντας το παράδειγμα I-L38>BY14026. – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT
Pingback: I-M170 and the Babylonian confusion of Y-Haplogroup names – ΑΝΤΩΝΙΟΣ ΔΝΑ PROJECT